Setup of a shared folder in a home cluster
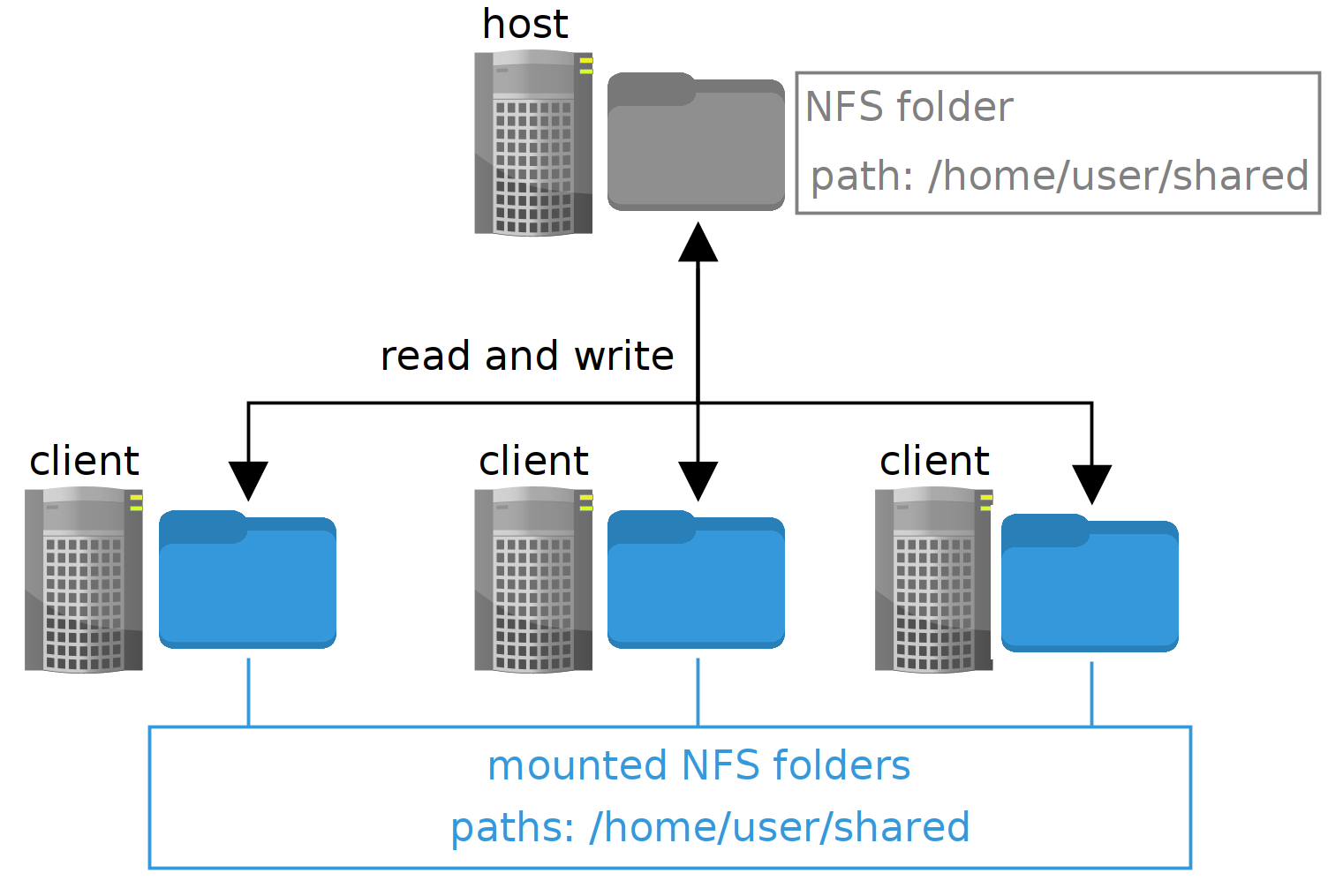 Image credit: Blas M. Benito
Image credit: Blas M. Benito
In the previous posts I have covered how to setup a home cluster, and how to run parallel processes with foreach in R. However, so far I haven’t covered how to setup a folder shared among the cluster nodes to store the results of parallel computations.
This post (developed on Ubuntu and Ubuntu Server 20.04) assumes that the home cluster runs on Linux, and covers the following topics.
- Basics of the Network File System protocol (NFS).
- Setup of an NFS folder in a home cluster.
- Using an NFS folder in a parallelized loop.
The Network File System protocol (NFS)
The Network File System protocol offers the means for a host computer to allow other computers in the network (clients) to read and write the contents of a given folder. The clients access such folder like if it was a local one, while in fact it is a reference to the one in the host computer.
The image at the beginning of the post illustrates the concept. There is a host computer with a folder in the path /home/user/cluster_shared (were user is your user name) that is broadcasted to the network, and there are one or several clients that are mounting mounting (making accessible) the same folder in their local paths /home/user/cluster_shared.
If the host writes a file to the shared folder, it is available right away for the clients, and the other way around. At the end, the idea is to have a folder shared among all computers in the cluster, while having the same exact path on each one of them to write or read files from such shared folder.
Setup of an NFS folder in a home cluster
To setup the shared folder we’ll need to do some things in the host, and some things in the clients. Let’s start with the host.
Preparing the host computer
First we need to install the nfs-kernel-server.
sudo apt update
sudo apt install nfs-kernel-serverNow we can create the shared folder. Remember to replace user with your user name, and cluster_shared with the actual folder name you want to use.
mkdir /home/user/cluster_sharedTo broadcast it we need to open the file /etc/exports…
sudo gedit /etc/exports… and add the following line
/home/user/cluster_shared IP_CLIENT1(rw,no_subtree_check) IP_CLIENT2(rw,no_subtree_check) IP_CLIENT3(rw,no_subtree_check)where:
/home/user/cluster_sharedis the path of the shared folder.IP_CLIENTxare the IPs of each one of the clients.rwgives reading and writing permission on the shared folder to the given client.no_subtree_checkprevents the host from checking the complete tree of shares before attending a request (read or write) by a client.
For example, the last line of my /etc/exports file looks like this:
/home/blas/cluster_shared 10.42.0.34(rw,async,no_subtree_check) 10.42.0.104(rw,async,no_subtree_check)Save the file, and to make the changes effective, execute:
sudo exportfs -raTo end preparing the host we have to update the firewall rules to allow nfs connections from the clients. Notice that one rule per client needs to be defined, using the clients IPs to identify them.
sudo ufw allow from IP_CLIENT1 to any port nfs
sudo ufw allow from IP_CLIENT2 to any port nfs
sudo ufw statusPreparing the clients
First we have to install the Linux package nfs-common on each client.
sudo apt update
sudp apt install nfs-commonNow we can create a folder in the clients and use it to mount the NFS folder of the host.
mkdir -p /home/user/cluster_shared
sudo mount IP_HOST:/home/user/cluster_shared /home/user/cluster_sharedThe second line of code is mounting the folder /home/user/cluster_shared of the host in the folder /home/user/cluster_shared of the client.
To make the mount permanent, we have to open /etc/fstab with super-user privilege in the clients…
sudo gedit /etc/fstab… and add the line
IP_HOST:/home/user/cluster_shared /home/user/cluster_shared nfs defaults 0 0Remember to replace IP_HOST and user with the right values!
Now we can test that the shared folder works as intended by writing one file into it from each computer in the network using the code below in the command line.
cd cluster_shared
touch filename.txt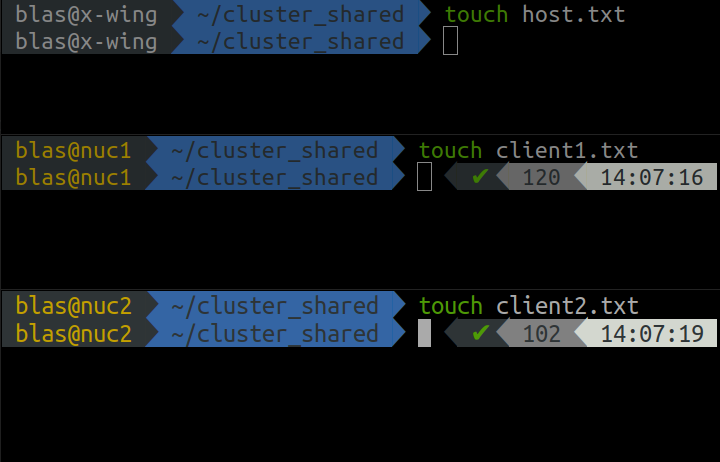
Once the files are created, we can check they are visible from each computer using the ls command.
ls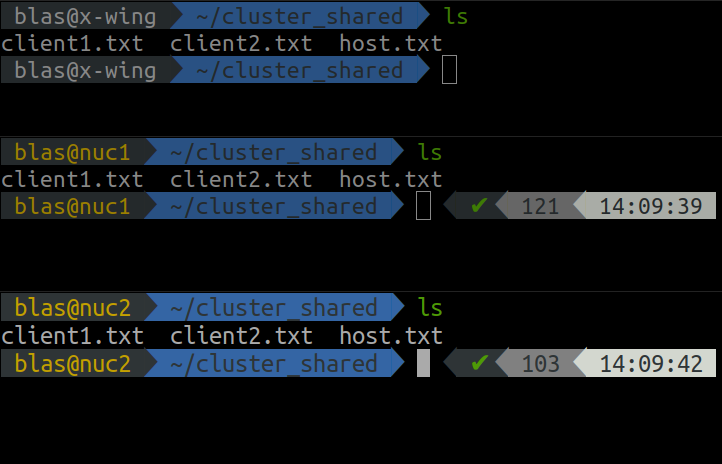
Using an NFS folder in a parallelized loop
In a previous post I described how to run parallelized tasks with foreach in R. This section intends to complete that post by showing how a shared NFS folder can be used to store input and output files during the execution of a parallelized loop
The task
In this hypothetical example we have a large number of data frames stored in /home/user/cluster_shared/input. Each data frame has the same predictors a, b, c, and d, and a different response variable, named y1 for the data frame y1, y2 for the data frame y2, and so on. In the example we will be using 100 data frames, but there could be thousands. This example would be valid as well for any other kind of object, such as raster files, time-series, etc etc.
First we have to load the libraries we’ll be using.
#automatic install of packages if they are not installed already
list.of.packages <- c(
"foreach",
"doParallel",
"ranger"
)
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages) > 0){
install.packages(new.packages, dep=TRUE)
}
#loading packages
for(package.i in list.of.packages){
suppressPackageStartupMessages(
library(
package.i,
character.only = TRUE
)
)
}The code chunk below generates the folder /home/user/cluster_shared/input and populates it with the dummy files.
#creating the input folder
input.folder <- "/home/blas/cluster_shared/input"
dir.create(input.folder)
#data frame names
df.names <- paste0("y", 1:100)
#filling it with files
for(i in df.names){
#creating the df
df.i <- data.frame(
y = rnorm(1000),
a = rnorm(1000),
b = rnorm(1000),
c = rnorm(1000),
d = rnorm(1000)
)
#changing name of the response variable
colnames(df.i)[1] <- i
#assign to a variable with name i
assign(i, df.i)
#saving the object
save(
list = i,
file = paste0(input.folder, "/", i, ".RData")
)
#removing the generated data frame form the environment
rm(list = i, df.i, i)
}Our target now will be to fit one ranger::ranger() model per data frame stored in /home/blas/cluster_shared/input, save the model result to a folder with the path /home/blas/cluster_shared/input, and write a small summary of the model to the output of foreach.
Such target is based on this rationale: When executing a foreach loop as in x <- foreach(...) %dopar% {...}, the variable x is going to grow in memory very fast, competing for RAM resources with the worker nodes. Furthermore, since x is being written on the fly, the results would be lost if the computer crashes. When the size of the input and the output of our parallelized operation is larger than memory, we can use an NFS folder to store inputs and outputs, while keeping the RAM memory free for computational tasks only, with the positive side effect of having our outputs already stored should our computer decide to crash.
Also, please notice that here I am focusing in a cluster setting, but using a folder to read and write data during a loop paralellized with foreach can indeed be done in a single computer without an NFS folder. Any folder in your system will do the trick as well!
So, from here, we are going to prepare the cluster, and execute a parallelized loop fitting one model per data frame that reads the inputs and writes the outputs to the shared folder.
Cluster setup
We will also need the function I showed in the previous post to generate the cluster specification from a GitHub Gist.
source("https://gist.githubusercontent.com/BlasBenito/93ee54d3a98d101754aaff0d658dccca/raw/de57b23740ca90bc02fbd0d5cd3551106ff2fb6d/cluster_spec.R")Below I use the function to create a cluster specification and initiate the cluster with parallel::makeCluster().
#generate cluster specification
spec <- cluster_spec(
ips = c('10.42.0.1', '10.42.0.34', '10.42.0.104'),
cores = c(7, 4, 4),
user = "blas"
)
#define parallel port
Sys.setenv(R_PARALLEL_PORT = 11000)
Sys.getenv("R_PARALLEL_PORT")
#setting up cluster
my.cluster <- parallel::makeCluster(
master = '10.42.0.1',
spec = spec,
port = Sys.getenv("R_PARALLEL_PORT"),
outfile = "",
homogeneous = TRUE
)
#check cluster definition (optional)
print(my.cluster)
#register cluster
doParallel::registerDoParallel(cl = my.cluster)
#check number of workers
foreach::getDoParWorkers()Parallelized loop
For everything to work as intended, we first need to create the output folder.
output.folder <- "/home/blas/cluster_shared/output"
dir.create(output.folder)And now we are ready to execute the parallelized loop. Notice that I am using the output of list.files() to have a vector of file names the loop will iterate over. The steps inside of the loop go as follows:
- 1. Remove the extension
.RDatafrom the file name. We’ll later use the result to useassign()on the fitted model to change its name to the same as the input file before saving it. - 2. Read the input data frame and store in an object named
df. - 3. Fit the model with ranger, using the first column of
dfas respose variable. - 4. Change the model name to the name of the input file without extension, resulting from the first step described above.
- 5. Save the model into the output folder with the extension
.RData. - 6. Return a data frame with one line with the name of the response variable, the r-squared, and the importance of each predictor.
#list of input files as iterator
input.files <- list.files(
path = input.folder,
full.names = FALSE
)
modelling.summary <- foreach(
input.file = input.files,
.combine = 'rbind',
.packages = "ranger"
) %dopar% {
# 1. input file name without extension
input.file.name <- tools::file_path_sans_ext(input.file)
# 2. read input file
df <- get(load(paste0(input.folder, "/", input.file)))
# 3. fit model
m.i <- ranger::ranger(
data = df,
dependent.variable.name = colnames(df)[1],
importance = "permutation"
)
# 4. change name of the model to one of the response variable
assign(input.file.name, m.i)
# 5. save model
save(
list = input.file.name,
file = paste0(output.folder, "/", input.file)
)
# 6. returning summary
return(
data.frame(
response.variable = input.file.name,
r.squared = m.i$r.squared,
importance.a = m.i$variable.importance["a"],
importance.b = m.i$variable.importance["b"],
importance.c = m.i$variable.importance["c"],
importance.d = m.i$variable.importance["d"]
)
)
}Once this parallelized loop is executed, the folder /home/blas/cluster_shared/output should be filled with the results from the cluster workers, and the modelling.summary data frame contains the summary of each fitted model.
 Now that the work is done, we can stop the cluster.
Now that the work is done, we can stop the cluster.
parallel::stopCluster(cl = my.cluster)Now you know how to work with data larger than memory in a parallelized loop!